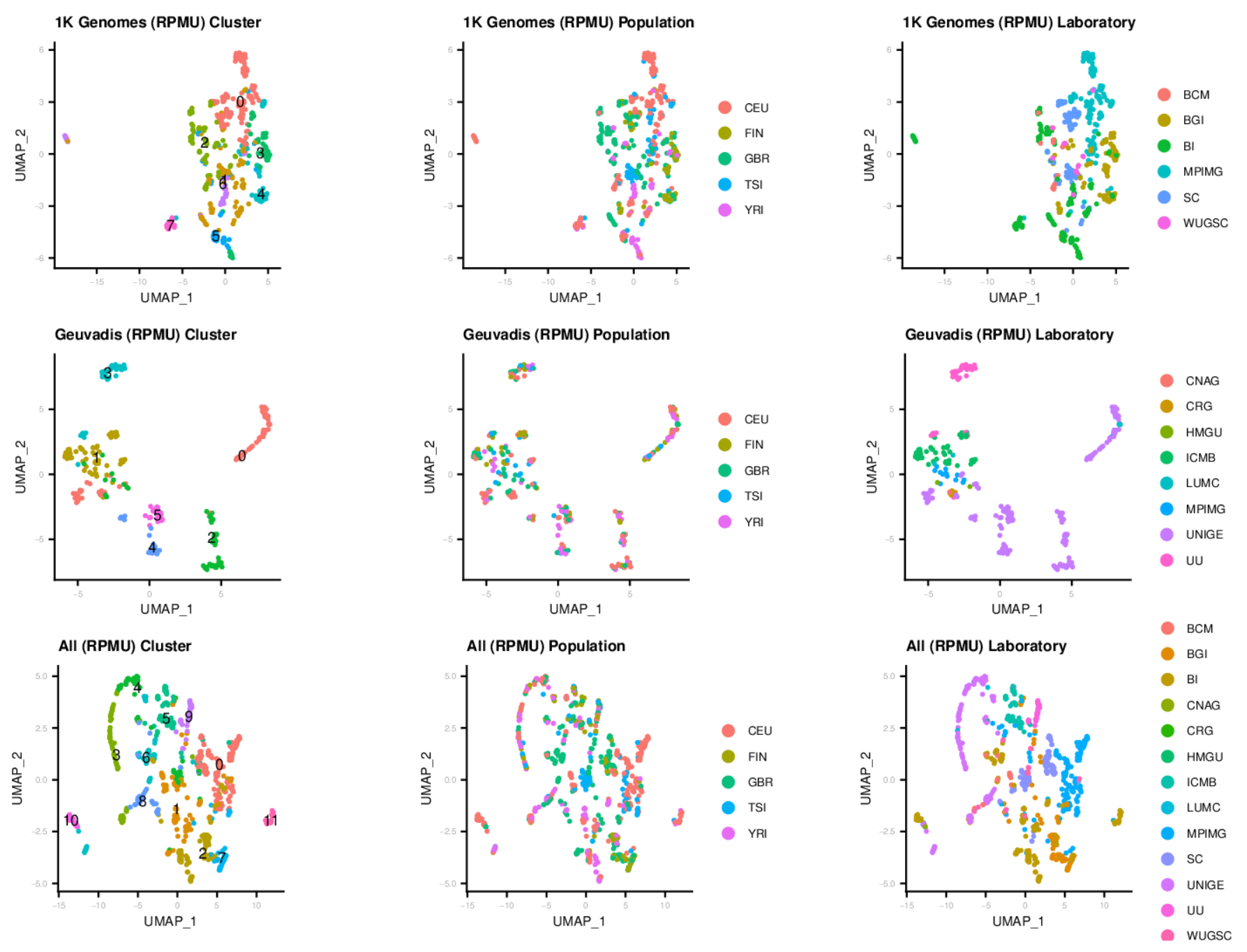
Clustering of sequencing runs with contamination profiles
After profiling the contamination of 1K Genomes and Geuvadis samples by conducting the OpenContami pipeline, we run Seurat3 for clustering them. At here, we excluded LCV (lymphocryptovirus; HHV4) and PhiX174microvirus (Illumina spike-in). The figures show that the runs were separated by sequencing laboratories more preferentially. Importantly, the sequencing protocols (i.e. DNA-seq or RNA-seq) clearly separated the runs. These results suggest that the presence of contaminants is associated with the laboratory environment and the contamination profiles are different among sequencing protocols.
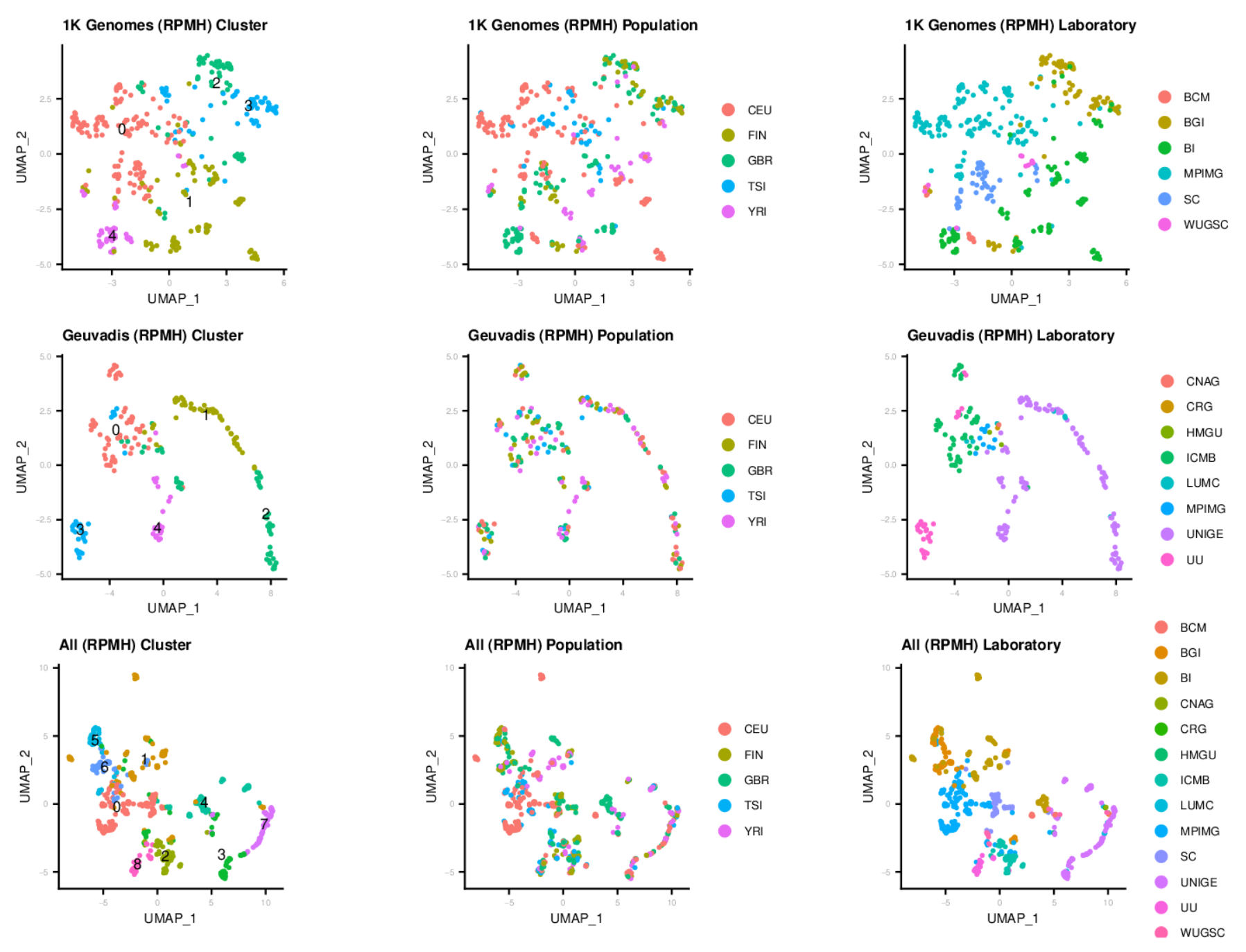
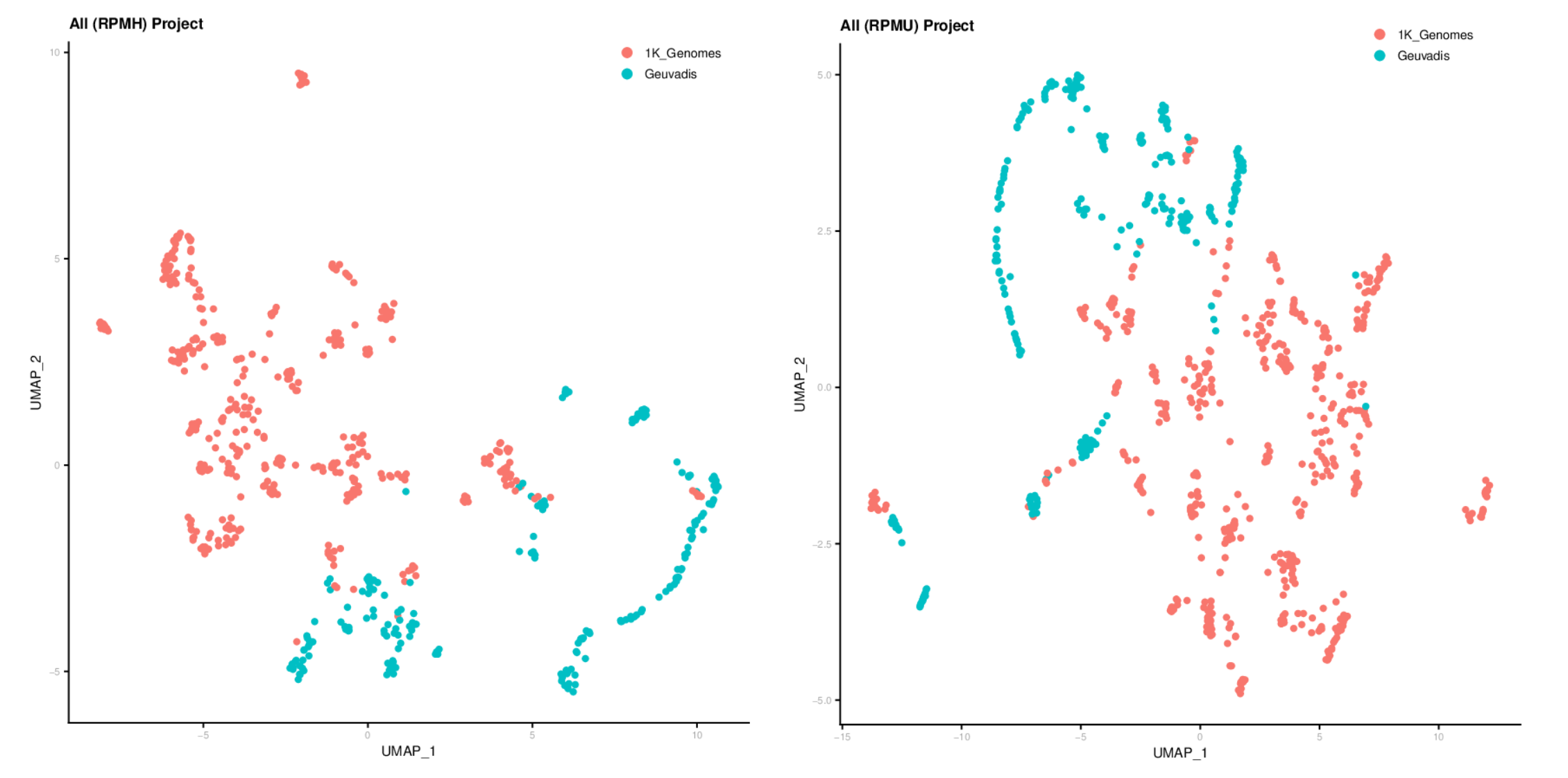
After profiling the contamination of 1K Genomes and Geuvadis samples by conducting the OpenContami pipeline, we run Seurat3 for clustering them. At here, we excluded LCV (lymphocryptovirus; HHV4) and PhiX174microvirus (Illumina spike-in). The figures show that the runs were separated by sequencing laboratories more preferentially. Importantly, the sequencing protocols (i.e. DNA-seq or RNA-seq) clearly separated the runs. These results suggest that the presence of contaminants is associated with the laboratory environment and the contamination profiles are different among sequencing protocols.